Examples
Test Cases
The sciform
test suite
contains hundreds of example formatting test cases which showcase the
many available formatting options.
Formatter
Here are a small selection of examples which demonstrate some of the available formatting options.
>>> from sciform import Formatter
>>> num = 12345.54321
>>> formatter = Formatter(exp_mode="scientific", round_mode="sig_fig", ndigits=4)
>>> print(formatter(num))
1.235e+04
>>> formatter = Formatter(
... exp_mode="engineering",
... round_mode="dec_place",
... ndigits=10,
... sign_mode=" ",
... superscript=True,
... )
>>> print(formatter(num))
12.3455432100×10³
>>> formatter = Formatter(
... exp_mode="fixed_point",
... upper_separator=" ",
... decimal_separator=",",
... lower_separator="_",
... sign_mode="+",
... )
>>> print(formatter(num))
+12 345,543_21
>>> num = 0.076543
>>> formatter = Formatter(
... exp_mode="scientific", exp_val=-3, exp_format="parts_per", add_ppth_form=True
... )
>>> print(formatter(num))
76.543 ppth
>>> formatter = Formatter(
... exp_mode="scientific", exp_val=-2, exp_format="prefix", add_c_prefix=True
... )
>>> print(formatter(num))
7.6543 c
>>> formatter = Formatter(exp_mode="scientific", exp_val=-6, exp_format="prefix")
>>> print(formatter(num))
76543 μ
>>> formatter = Formatter(exp_mode="percent")
>>> print(formatter(num))
7.6543%
>>> num = 3141592.7
>>> unc = 1618
>>> formatter = Formatter()
>>> print(formatter(num, unc))
3141593 ± 1618
>>> formatter = Formatter(
... exp_mode="engineering",
... exp_format="prefix",
... pdg_sig_figs=True,
... pm_whitespace=False,
... )
>>> print(formatter(num, unc))
(3.1416±0.0016) M
>>> num = 314159.27
>>> unc = 1618
>>> formatter = Formatter(
... exp_mode="engineering_shifted", pdg_sig_figs=True, paren_uncertainty=True
... )
>>> print(formatter(num, unc))
0.3142(16)e+06
Format Specification Mini Language
>>> from sciform import SciNum
>>> print(f"{SciNum(1234.432):0=+5.6f}")
+001234.432000
In the preceding example 0= indicates that left padding should be
done using '0' characters.
The + indicates that a sign symbol should always be displayed.
The 5 preceding the round mode symbol (.) indicates that the
number should be left padded to the 10+5 (ten-thousands)
decimal place.
The .6 indicates that the number will be rounded to six digits past
the decimal points.
The f indicates that the number will be displayed in fixed point
exponent mode.
>>> print(f"{SciNum(123, 0.123):#!2R()}")
0.12300(12)E+03
In the preceding example the # alternate flag combined with R
indicates that the number will be formatted in the shifted engineering
exponent mode with a capitalized exponent symbol 'E'.
!2 indicates that the number will be rounded so that the uncertainty
has two significant figures.
The () indicates that the value/uncertainty pair should be formatted
using the parentheses uncertainty format.
>>> print(f"{SciNum(123):ex-3p}")
123000 m
In the preceding example the e indicates that scientific notation
should be used.
The x-3 indicates that the exponent will be forced to equal -3.
Finally the p indicates that the SI prefix mode should be used.
>>> print(f"{SciNum(123): .-1f}")
120
In this example the leading space indicates a leading space should be
included for non-negative numbers so that positive and negative numbers
have the same justification.
The .-1 indicates the number should be rounded to one
digit before the decimal point and the f indicates that fixed
point mode should be used.
SciNum, and Global Options
Here are a small selection of examples which demonstrate some of the
available string formatting options.
Note that many options are not available through the Format Specification Mini-Language, so
these options must be selected by configuring the global options.
Here this is done using the GlobalOptionsContext context
manager, but this could have been done using set_global_options()
instead.
>>> from sciform import SciNum, GlobalOptionsContext
>>> num = SciNum(12345.54321)
>>> print(f"{num:!4e}")
1.235e+04
>>> print(f"{num: .10r}")
12.3455432100e+03
>>> with GlobalOptionsContext(
... upper_separator=" ",
... decimal_separator=",",
... lower_separator="_",
... ):
... print(f"{num:+}")
+12 345,543_21
>>> num = SciNum(0.076543)
>>> with GlobalOptionsContext(exp_format="parts_per", add_ppth_form=True):
... print(f"{num:ex-3}")
...
76.543 ppth
>>> with GlobalOptionsContext(exp_format="prefix", add_c_prefix=True):
... print(f"{num:ex-2}")
...
7.6543 c
>>> with GlobalOptionsContext(exp_mode="scientific", exp_val=-6, exp_format="prefix"):
... print(f"{num:ex-6}")
...
76543 μ
>>> print(f"{num:%}")
7.6543%
>>> num_unc = SciNum(3141592.7, 1618)
>>> print(f"{num_unc}")
3141593 ± 1618
>>> with GlobalOptionsContext(pdg_sig_figs=True, pm_whitespace=False):
... print(f"{num_unc:rp}")
...
(3.1416±0.0016) M
>>> num_unc = SciNum(314159.27, 1618)
>>> with GlobalOptionsContext(pdg_sig_figs=True):
... print(f"{num_unc:#r()}")
...
0.3142(16)e+06
Plotting and Tabulating Fit Data
We are given 3 data sets:
Data
{
"data_0": {
"x": [
-0.0001,
-7.777777777777778e-05,
-5.555555555555556e-05,
-3.3333333333333335e-05,
-1.1111111111111112e-05,
1.1111111111111112e-05,
3.3333333333333335e-05,
5.555555555555556e-05,
7.777777777777778e-05,
0.0001
],
"y": [
1000028705.1993496,
1000032562.3072652,
1000001896.3076739,
1000015911.7068863,
1000010552.1178038,
1000024250.5937256,
1000078382.654146,
1000099609.1405739,
1000156104.3810261,
1000218228.6977944
],
"y_err": [
10074.736165669336,
8935.559150444922,
9783.419334631266,
9603.772217328124,
8958.492409007613,
9691.25932834366,
9465.18285498962,
9252.428221037011,
10506.00188280341,
9674.67988710319
]
},
"data_1": {
"x": [
-0.0001,
-7.777777777777778e-05,
-5.555555555555556e-05,
-3.3333333333333335e-05,
-1.1111111111111112e-05,
1.1111111111111112e-05,
3.3333333333333335e-05,
5.555555555555556e-05,
7.777777777777778e-05,
0.0001
],
"y": [
1000101738.923473,
1000075626.403057,
1000047095.4124904,
1000035342.0517923,
1000034627.8667482,
1000024117.4097912,
1000032427.2038687,
1000058361.2708515,
1000090132.8138337,
1000137137.590938
],
"y_err": [
9546.933912908422,
9865.10610577111,
11233.801341794,
8709.471433431938,
9260.554239646424,
11037.621922605267,
11397.162303260564,
10037.634586482105,
10076.884695349665,
9877.999777816845
]
},
"data_2": {
"x": [
-0.0001,
-7.777777777777778e-05,
-5.555555555555556e-05,
-3.3333333333333335e-05,
-1.1111111111111112e-05,
1.1111111111111112e-05,
3.3333333333333335e-05,
5.555555555555556e-05,
7.777777777777778e-05,
0.0001
],
"y": [
1000162524.5101302,
1000112400.2588283,
1000095882.1735442,
1000055167.7660033,
1000026965.91884,
1000019054.4158406,
1000025166.114796,
1000035728.0662737,
1000075468.031305,
1000105069.4047513
],
"y_err": [
10476.46500331082,
10086.821923665966,
9559.819918015002,
10775.7594031851,
11296.833796893989,
10205.878907138671,
10025.395431977211,
10091.52840469254,
11880.794221192386,
10936.463066427263
]
}
}
We want to perform quadratic fits to these data sets, visualize
the results, and print the best fit parameters including the uncertainty
reported by the fit routine.
For these tasks we will require the numpy, scipy,
matplotlib, and tabulate packages.
Without sciform
Without sciform we can perform the fit and plot the data and best
fit lines and print out a table of best fit parameters and
uncertainties:
Code
from __future__ import annotations
import json
from pathlib import Path
from typing import TYPE_CHECKING
import matplotlib.pyplot as plt
import numpy as np
from scipy.optimize import curve_fit
from tabulate import tabulate
if TYPE_CHECKING:
from numpy.typing import NDArray
def quadratic(x: NDArray, c: float, x0: float, y0: float) -> NDArray:
return (c / 2) * (x - x0) ** 2 + y0
def main() -> None:
data_path = Path("data", "fit_data.json")
with data_path.open() as f:
data_dict = json.load(f)
color_list = ["red", "blue", "purple"]
fit_results_list = []
fig, ax = plt.subplots(1, 1)
for idx, single_data_dict in enumerate(data_dict.values()):
x = single_data_dict["x"]
y = single_data_dict["y"]
y_err = single_data_dict["y_err"]
fit_results_dict = {}
color = color_list[idx]
ax.errorbar(x, y, y_err, marker="o", linestyle="none", color=color, label=color)
# noinspection PyTupleAssignmentBalance
popt, pcov = curve_fit(quadratic, x, y, sigma=y_err, p0=(2e13, 0, 1e9))
model_x = np.linspace(min(x), max(x), 100)
model_y = quadratic(model_x, *popt)
ax.plot(model_x, model_y, color=color)
fit_results_dict["color"] = color
fit_results_dict["curvature"] = popt[0]
fit_results_dict["curvature_err"] = np.sqrt(pcov[0, 0])
fit_results_dict["x0"] = popt[1]
fit_results_dict["x0_err"] = np.sqrt(pcov[1, 1])
fit_results_dict["y0"] = popt[2]
fit_results_dict["y0_err"] = np.sqrt(pcov[2, 2])
fit_results_list.append(fit_results_dict)
ax.grid(True) # noqa: FBT003
ax.legend()
fig.savefig("outputs/fit_plot_no_sciform.png", facecolor="white")
plt.show()
table_str = tabulate(
fit_results_list,
tablefmt="grid",
headers="keys",
floatfmt="#.2g",
)
table_path = Path("outputs", "fit_plot_no_sciform_table.txt")
with table_path.open("w") as f:
f.write(table_str)
print(table_str) # noqa: T201
if __name__ == "__main__":
main()
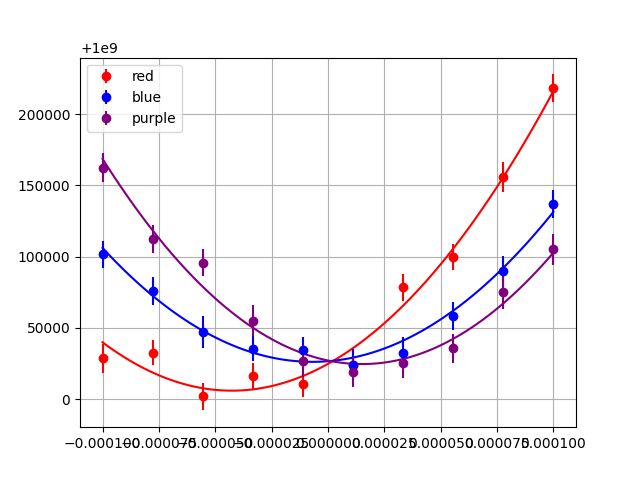
This produces the plot:
And the table:
+---------+-------------+-----------------+----------+----------+---------+----------+
| color | curvature | curvature_err | x0 | x0_err | y0 | y0_err |
+=========+=============+=================+==========+==========+=========+==========+
| red | 2.1e+13 | 1.9e+12 | -4.3e-05 | 4.8e-06 | 1.0e+09 | 4.6e+03 |
+---------+-------------+-----------------+----------+----------+---------+----------+
| blue | 1.8e+13 | 1.0e+12 | -6.9e-06 | 1.6e-06 | 1.0e+09 | 2.9e+03 |
+---------+-------------+-----------------+----------+----------+---------+----------+
| purple | 2.2e+13 | 1.7e+12 | 1.5e-05 | 2.5e-06 | 1.0e+09 | 4.4e+03 |
+---------+-------------+-----------------+----------+----------+---------+----------+
This plot and table suffer from a number of shortcomings which impede human readability.
In the table, the exponents for the values and uncertainties differ, making it hard to identify the significant digits of the value.
The number of digits displayed for the values is not correlated with the uncertainty for that value. For example, the
y0values are shown with precision to the 10+8 place, but the uncertainty indicates precision down to the 10+3 place.In the table, the exponents vary from one dataset to the next. It is hard to see these differences at a glance.
The tick labels on the plot are illegible because each value has so many digits.
Of course, even without sciform, it would be possible to make
manual adjustments to the plot and the table to improve these data
visualizations.
However, sciform will allow us to make the required changes in a
general and automated way.
With sciform
We can address these problems using sciform by:
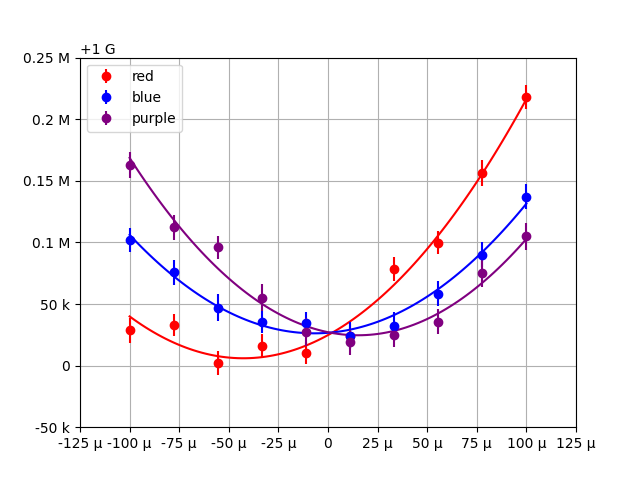
Using prefix scientific notation to label the plot axes. This will greatly reduce the number of symbols needed for each tick value.
Using value/uncertainty formatting in the table to collapse the value and error column pairs into individual columns. This will make obvious the relative scale between the uncertainty and the value. Using
sciform, the significant digits displayed for the value will always match the precision of the uncertainty. We will use the parentheses uncertainty format.Using engineering notation for the value/uncertainty in the table. This will make the relative scale between different rows obvious.
To do this we import sciform and make some helper functions for
displaying the plot axes as described:
Code
from __future__ import annotations
import json
import re
from pathlib import Path
from typing import TYPE_CHECKING, Literal
import matplotlib.pyplot as plt
import numpy as np
from sciform import Formatter
from scipy.optimize import curve_fit
from tabulate import tabulate
if TYPE_CHECKING:
from numpy.typing import NDArray
def get_scale_and_offset_from_offset_str(
ax: plt.Axes,
axis: Literal["x", "y"],
) -> tuple[float, float]:
"""Get the scale and offset for an axis formatted in scientific mode."""
plt.draw()
if axis == "x":
offset_text_obj = ax.xaxis.get_offset_text()
elif axis == "y":
offset_text_obj = ax.yaxis.get_offset_text()
else:
msg = f"axis must be 'x' or 'y', not '{axis}'."
raise ValueError(msg)
ax.ticklabel_format(axis=axis, style="sci")
ax.get_figure().canvas.draw() # Redraw canvas to update offset text
offset_text = offset_text_obj.get_text()
# Replace minus sign with hyphen minus sign
offset_text = offset_text.replace("\u2212", "-")
pattern = re.compile(r"^(?P<scale>1e[+-]?\d+)?(?P<offset>[+-]1e\d+)?$")
match = re.match(pattern, offset_text)
scale = float(match.group("scale") or 1)
offset = float(match.group("offset") or 0)
return scale, offset
def prefix_exp_ticks(
ax: plt.Axes,
axis: Literal["x", "y"],
*,
shifted: bool = False,
) -> None:
"""
Use prefix notation for axis tick labels. Scale the tick labels by
the multiplier that appears in the offset text and format the labels
into SI prefix format. Format any remaining offset value in the
offset text into SI prefix format as well.
"""
if not shifted:
exp_mode = "engineering"
else:
exp_mode = "engineering_shifted"
# noinspection PyTypeChecker
tick_formatter = Formatter(exp_mode=exp_mode, exp_format="prefix")
# noinspection PyTypeChecker
offset_formatter = Formatter(sign_mode="+", exp_mode=exp_mode, exp_format="prefix")
ax.ticklabel_format(axis=axis, style="sci")
if axis == "x":
old_ticklabels = ax.get_xticklabels()
elif axis == "y":
old_ticklabels = ax.get_yticklabels()
else:
msg = f"axis must be 'x' or 'y', not '{axis}'."
raise ValueError(msg)
scale, offset = get_scale_and_offset_from_offset_str(ax, axis)
new_tick_locations = []
new_tick_labels = []
for old_ticklabel in old_ticklabels:
x, y = old_ticklabel.get_position()
if axis == "x":
new_tick_locations.append(x)
else:
new_tick_locations.append(y)
# Replace minus sign with hyphen minus sign
old_label_str = old_ticklabel.get_text().replace("\u2212", "-")
val = float(old_label_str) * scale
new_str = tick_formatter(val)
new_tick_labels.append(new_str)
if offset != 0:
offset_str = offset_formatter(offset)
else:
offset_str = ""
if axis == "x":
ax.set_xticks(new_tick_locations, new_tick_labels)
ax.text(x=1.01, y=0, s=offset_str, transform=ax.transAxes)
else:
ax.set_yticks(new_tick_locations, new_tick_labels)
ax.text(x=0, y=1.01, s=offset_str, transform=ax.transAxes)
def quadratic(x: NDArray, c: float, x0: float, y0: float) -> NDArray:
return (c / 2) * (x - x0) ** 2 + y0
def main() -> None:
fit_results_formatter = Formatter(
exp_mode="engineering",
round_mode="sig_fig",
paren_uncertainty=True,
ndigits=2,
)
data_path = Path("data", "fit_data.json")
with data_path.open() as f:
data_dict = json.load(f)
color_list = ["red", "blue", "purple"]
fit_results_list = []
fig, ax = plt.subplots(1, 1)
for idx, single_data_dict in enumerate(data_dict.values()):
x = single_data_dict["x"]
y = single_data_dict["y"]
y_err = single_data_dict["y_err"]
fit_results_dict = {}
color = color_list[idx]
ax.errorbar(x, y, y_err, marker="o", linestyle="none", color=color, label=color)
# noinspection PyTupleAssignmentBalance
popt, pcov = curve_fit(quadratic, x, y, sigma=y_err, p0=(2e13, 0, 1e9))
model_x = np.linspace(min(x), max(x), 100)
model_y = quadratic(model_x, *popt)
ax.plot(model_x, model_y, color=color)
fit_results_dict["color"] = color
fit_results_dict["curvature"] = fit_results_formatter(
popt[0],
np.sqrt(pcov[0, 0]),
)
fit_results_dict["x0"] = fit_results_formatter(popt[1], np.sqrt(pcov[1, 1]))
fit_results_dict["y0"] = fit_results_formatter(popt[2], np.sqrt(pcov[2, 2]))
fit_results_list.append(fit_results_dict)
ax.grid(True) # noqa: FBT003
ax.legend()
prefix_exp_ticks(ax, "x")
prefix_exp_ticks(ax, "y", shifted=True)
fig.savefig("outputs/fit_plot_with_sciform.png", facecolor="white")
plt.show()
table_str = tabulate(fit_results_list, headers="keys", tablefmt="grid")
table_path = Path("outputs", "fit_plot_with_sciform_table.txt")
with table_path.open("w") as f:
f.write(table_str)
print(table_str) # noqa: T201
if __name__ == "__main__":
main()
This produces the plot:
and the table:
+---------+-----------------+------------------+---------------------+
| color | curvature | x0 | y0 |
+=========+=================+==================+=====================+
| red | (20.7(1.9))e+12 | (-42.7(4.8))e-06 | (1.0000060(46))e+09 |
+---------+-----------------+------------------+---------------------+
| blue | (18.4(1.0))e+12 | (-6.9(1.6))e-06 | (1.0000262(29))e+09 |
+---------+-----------------+------------------+---------------------+
| purple | (21.7(1.7))e+12 | (15.1(2.5))e-06 | (1.0000246(44))e+09 |
+---------+-----------------+------------------+---------------------+
We can see the plot and table are immediately much more legible. Less characters are needed to communicate the data in both visualizations. The relative scaling of parameters between datasets and the relative scaling between the value and uncertainty for each entry are immediately clear.
Mapping Over Collections
It may be convenient to apply sciform mapping to collections (sets,
lists, arrays, etc.) of numbers.
>>> from sciform import Formatter
>>>
>>> formatter = Formatter(
... exp_mode="engineering",
... exp_format="prefix",
... pdg_sig_figs=True,
... paren_uncertainty=True,
... )
>>> val_list = [1000, 2000, 3000]
>>> err_list = [200, 400, 600]
>>>
>>> val_str_list = list(map(formatter, val_list))
>>> print(val_str_list)
['1 k', '2 k', '3 k']
>>>
>>> val_err_str_list = list(map(formatter, val_list, err_list))
>>> print(val_err_str_list)
['1.00(20) k', '2.0(4) k', '3.0(6) k']
We may also want to map over higher dimensional numpy arrays. numpy.vectorize() makes this easy.
>>> import numpy as np
>>>
>>> vec_formatter = np.vectorize(formatter)
>>> arr = np.array([[1e6, 2e6, 3e6], [4e6, 5e6, 6e6], [7e6, 8e6, 9e6]])
>>>
>>> arr_err = np.array([[9e4, 8e4, 7e4], [6e4, 5e4, 4e4], [3e4, 2e4, 1e4]])
>>>
>>> print(vec_formatter(arr))
[['1 M' '2 M' '3 M']
['4 M' '5 M' '6 M']
['7 M' '8 M' '9 M']]
>>> print(vec_formatter(arr, arr_err))
[['1.00(9) M' '2.00(8) M' '3.00(7) M']
['4.00(6) M' '5.00(5) M' '6.00(4) M']
['7.000(30) M' '8.000(20) M' '9.000(10) M']]